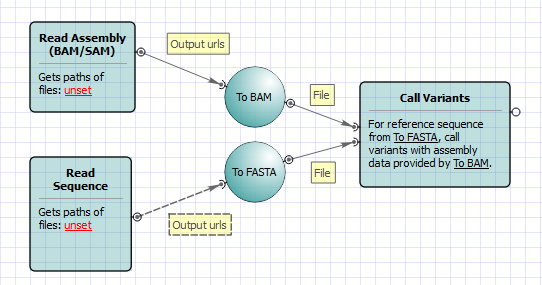
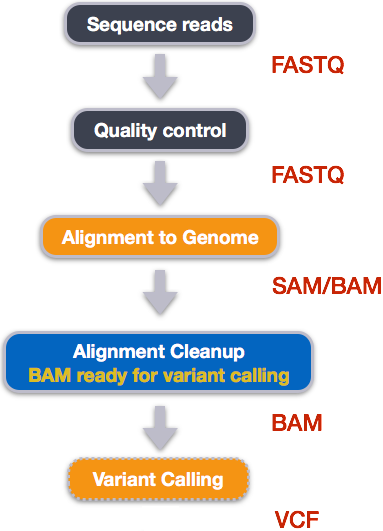
GitHub - samtools/bcftools: This is the official development repository for BCFtools. See installation instructions and other documentation here http://samtools.github.io/bcftools/howtos/install.html
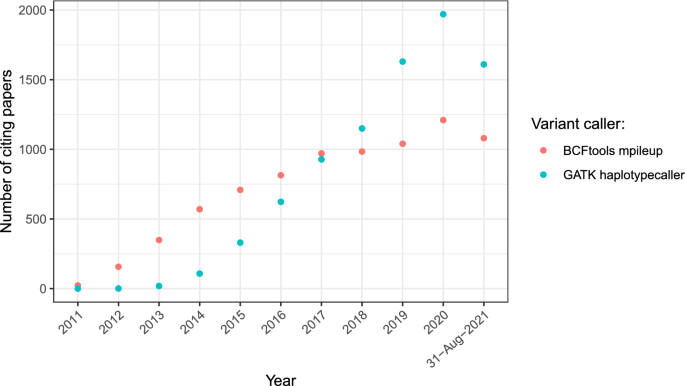
The evaluation of Bcftools mpileup and GATK HaplotypeCaller for variant calling in non-human species | Scientific Reports
![PDF] BCFtools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data | Semantic Scholar PDF] BCFtools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/9855a97fa2a760c4bfe74f5902f9de458f0a5a79/2-Figure1-1.png)
PDF] BCFtools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data | Semantic Scholar

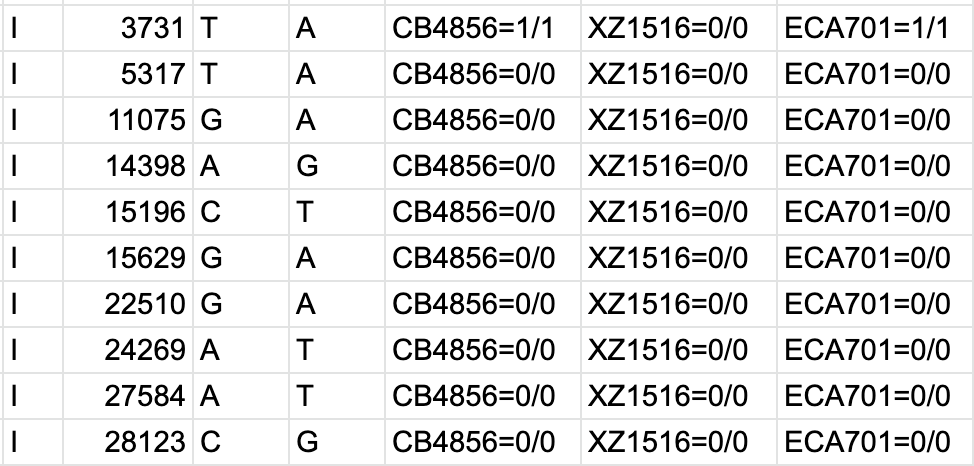
bcftools mpileup - Difference between IDV and FORMAT/AD* fields · Issue #912 · samtools/bcftools · GitHub